About Brainhack Warsaw 2019
On the first weekend of March 2019, the second edition of Brainhack Warsaw will take place. During this three-day event dedicated to students and PhD students, we will work in teams on neuroscience-related projects.
The aim of the event is to meet new, enthusiastic researchers, make new friendships in academia, learn, share the knowledge on data mining and brain research, but also promote open science in the spirit of the whole Brainhack community (Craddock et al., 2016). Attendees of various backgrounds are welcome to join!
By submitting your own, genuine research project, you can gain a priceless leadership experience as you will manage a group of researchers at our three-day event.
Please note Brainhack Warsaw 2019 is a sister event for the interdisciplinary Aspects of Neuroscience conference which will take place on 23-25th November 2018 in Warsaw. Participation in the conference is not mandatory to take part in Brainhack but we encourage to also consider this.
Deadline for project proposals: 11.01.2019
Announcement of projects: 20.01.2019
Participant registration starts: 21.01.2019
Deadline for participant registration: 24.02.2019
Please send all the related questions at the mailing address: brainhackwarsaw@gmail.com
Brainhack Warsaw - Past Editions
If you want to take a look at first edition: here you can view: Speakers, Projects and a lot of other interesting information about Brainhack Warsaw 2017.
Manifesto
At Brainhack Warsaw we believe that creativity is a key factor of success. We think that science, programming and art are similar in their nature. Art doesn’t care about hierarchy. Nor we do. There are paths yet to be discovered and boundaries yet to be transgressed. Brainhack Warsaw is open for everyone. People from academia, artists, data scientists, physicists, biologists, poets, psychologists, females, males, religious, atheists. There’s only one thing that matters: creativity.
That’s why we chose yellow as our new color. Yellow is a color of creativity. We know that it takes a lot of courage to think differently. We do not choose the easy way. We do not follow old trajectories. That’s what we are. That’s how we look. Join us.
Sincerely yours, ❤️
Brainhack Warsaw 2019 Team
Venue
Brainhack will take place at the University of Warsaw, Faculty of Physics, Pasteura 5, 02-093 Warsaw, Poland. Additional information how to get to Warsaw and university campus can be found at our sister-event’s site: Aspects of Neuroscience website.
Speakers
Towards a collaborative neuroscience
Abstract: Open Science is a recent movement aiming at giving researchers open access to datasets, tools, pipelines and publications. Science is no longer an ivory tower; it became more inclusive and transparent than ever before. How does research collaboration look like in times of open science? In this presentation, I will be talking about new tools that we are building to help researchers all around the globe work together. I will also relate to my daily research, including three main research lines in my group: brain development, evolution and Autism Spectrum Disorders.
Bio: Roberto Toro graduated from a Master’s degree in Universidad Técnica Federico Santa María, Valparaiso, Chile (1997) and Pierre and Marie Curie University, Paris (1998) and obtained a PhD from Pierre and Marie Curie University, Paris (2003). Today, he is a Group Leader at the Institut Pasteur, Paris. Roberto is an expert in the domain of development and evolution of brain anatomy. In his research group, he utilizes mathematical modelling, Magnetic Resonance Imaging and human genetics. The team is oriented at developing computational neuroanatomy methods to analyse the normal diversity of human brain anatomy, and find differences associated with neurodevelopmental pathologies. Roberto is particularly interested in Autism Spectrum Disorders. Roberto is also highly engaged in the open science movement. He was a cofounder of the Brainhack community (Craddock et al., 2016) and he is an awardee of the Open Science Prize 2016 (worth 80k USD) for the BrainBox project. He is also currently a member of the OHBM Open Science Special Interest Group, and the Co-Chair of the OHBM Hackathon.
Open mentoring
Abstract: One of the strongest movements in academia nowadays, is the Open Science movement. Its main postulates are: sharing access to high quality datasets with the general public and setting new standards for research reproducibility, e.g. by sharing codes and pipelines through open-access services such as GitHub. What else can and should we share in academia? Mentoring is one example of an asset which can also be shared between researchers all around the world. Today, it is no longer the case that mentoring experience can only be received from the direct supervisor; one can and should search out for advice from multiple researchers at every career stage, independently from geographical location - especially when there is little opportunity to be mentored in the local research environment.
Within the Organization for Human Brain Mapping Student and Postdoc Special Interest Group, we are interested in assisting early career researchers in accelerating their careers. One of our main initiatives is the International Online Mentoring Programme. Within the Programme, every member of the OHBM community worldwide can become either a mentor, a mentee or both. In this blitz talk, I will review the lessons we learned after over two years of running the programme, and recommendations we can give to young generation of researchers who are just entering their PhD.
Bio: Natalia Bielczyk is now completing her thesis within the Donders Graduate School, Donders Institute for Brain, Cognition and Behavior, Nijmegen, the Netherlands. Her research concerns developing new methods for connectomics in the domain of cognitive neuroimaging, i.e. for functional and effective connectivity research. She also currently holds a position of a Career Development and Mentoring Manager within the Organization for Human Brain Mapping Student and Postdoc Special Interest Group (www.ohbmtrainees.com).
BrainBox
Abstract: Progress in biomedical research requires studying datasets much larger than previously thought, but also finding ways for large numbers of researchers to collaborate creatively. In neuroimaging, a wealth of open data is available, including data on autism, schizophrenia, and many more. However, the data is not easy to find, is curated and analysed locally, in redundant, often wasteful and opaque ways. Our Web application BrainBox explores a radical open science solution to the problem of distributed collaboration. A mix of Wikipedia and Google Docs, BrainBox enables researchers to add a layer of real-time interactive annotation and segmentation to the great amount of neuroimaging data shared online. It allows distributed research teams to collaborate in the analysis of open data and avoid redundant work – promoting community efforts instead of competition. In my presentation, I will show you BrainBox in (inter)action, how to create a project, invite collaborators, annotate data, and use the API to integrate BrainBox into your automatic, reproducible analysis pipelines. Currently, more than 13,000 datasets are indexed and available for collaboration. During the Brainhack, I will be more than happy to help you set up your own projects, or invite you to join my current project. By working together, sharing our work and our analyses, we should improve transparency, increase statistical power and reproducibility.
Bio: I use neuroimaging to understand the evolution and development of the brain. I am strongly involved in open, reproducible, science and interdisciplinary research: I develop open Web applications to facilitate access to open data, foster collaboration and citizen science. I have a background in fine arts and in many of my projects I combine art and science (http://tiny.cc/ArtOfMR_2X) as a way to engage with the general public (http://tiny.cc/FireAndWireX).
International B2B communication (on a budget)
Abstract: Tips and tricks of building an international presence and business relations with various online tools. You’ll learn how to optimize your business profile, various networking strategies, and best practice of expert positioning. We’ll cover best practice in online communication on LinkedIn and Twitter, as well as how it supports offline interactions, e.g. during trade shows.
Bio: Agata is responsible for the communication strategy of Geek Girls Carrots. She’s a member of IEEE Global A/IS Ethics Initiative, HIMSS Interoperability & Health Information Exchange (HIE) Task Force and Digital Health Society coordinated by European Connected Health Alliance. She’s been working in international communication of tech brands for the past eight years. She graduated from Applied Linguistics at the Univerity of Warsaw and School of European and Spanish Law at Universidad Castilla-La Mancha/UW.
Projects
Project 1: Misfolded protein spreading on brain connectome
Project 2: Generating fake MRI to enhance Alzheimer’s disease prediction
Project 3: Network analysis of the electrical activity of the brain during epileptic seizures
Project 4: Deep Frankenstein: dissecting and sewing artificial neural networks
Project 5: Tackling the ABCD Neurocognitive Prediction Challenge
Project 6: Multidimensionality problem in passive BCI for game control
Project 8: EMG-based desktop controlling
Project 9: Mapping the state of modern psychology through the public online activity of scientists
Project 1: Misfolded protein spreading on brain connectome
UNAVAILABLE
Authors: Alessandro Crimi, PhD1 / Eleanna Kara 2
- University hospital of Zurcih, Switzerland
- Neuropathology resident at UniversitätsSpital Zürich, Switzerland
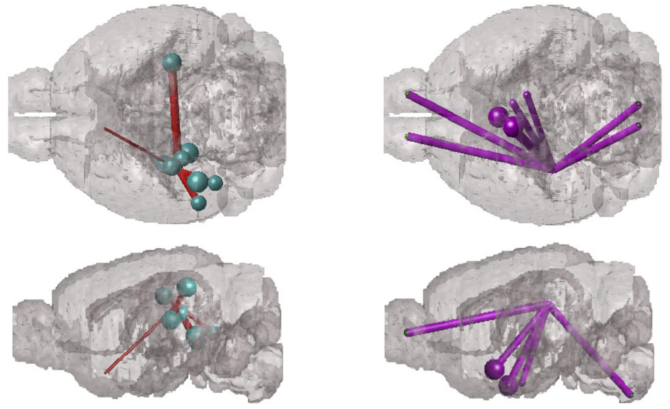
Abstract Connectomics have been used so far to look for quantifying global and local differences in the functional or structural brain networks [1], or alternatively, to simulate/study brain hemodynamics [2] . Very few studies have used connectomes to investigate the spreading of misfolded proteins which is at the basis of Parkinson’s (PD) and Alzheimer’s disease (AD) [3,4]. It is believed that diseases as AD and PD are spread by misfolded proteins or agents which moves along brain connections (axons and dendrides of the neurons) starting from specific regions to others [5]. For instance, AD has a progression of tau pathology consistently beginning in the entorhinal cortex, the locus coeruleus, and other nearby noradrenergic brainstem nuclei, before spreading to the rest of the limbic system as well as the cacingulate and retrosplenial cortices. While Parkinsons starts from the brainstem and spread to the neocortex [5]. A previous study investigated this mechanism on connectome comparing simulated tau deposits on connectome to those detected by PET scans specific for Alzheimer’s [3]. In this project, we aim at carrying out a similar study bu t for PD. In particular, we want to simulate deposits/spreading of alpha-syn proceeding via the brain’s anatomic connectivity network. We will use human and mice data provided by the supervisors. The main challenge of the project is defining a proper model of spreading along the connectome. Data from the PPMI dataset (https://www.ppmi-info.org/access-data-specimens/) and given by the project supervisors will be given. Those include case (PD patients) and control subjects along with clinical data and genetics.
A list of 1-5 key papers/materials summarising the subject:
- Griffa, Alessandra, et al. “Structural connectomics in brain diseases.” Neuroimage 80 (2013): 515-526. https://www.sciencedirect.com/science/article/pii/S1053811913004035
- Friston, Karl, Rosalyn Moran, and Anil K. Seth. “Analysing connectivity with Granger causality and dynamic causal modelling.” Current opinion in neurobiology 23.2 (2013): 172-178. https://www.sciencedirect.com/science/article/pii/S0959438812001845
- Iturria-Medina, Yasser, et al. “Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders.” PLoS computational biology 10.11 (2014): e1003956. https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003956
- Raj, Ashish, Amy Kuceyeski, and Michael Weiner. “A network diffusion model of disease progression in dementia.” Neuron73.6 (2012): 1204-1215. https://www.sciencedirect.com/science/article/pii/S0896627312001353
- H. Braak et al., “Staging of brain pathology related to sporadic Parkinsons disease,” Neurobiology of aging, vol. 24, no. 2, pp. 197–211, 2003. https://www.sciencedirect.com/science/article/pii/S0197458002000659
A list of requirements for taking part in the project:
- minimum level of English and basic concept of connectomics,
- some knowledge of neuropathology and dynamic models will be appreciated
- programming skills in Python, Matlab/Octave
A maximal number of participants: 15
Skills and competences you can learn during the project:
- Use connectomes to run simulations,
- Basic introduction on connectomics
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 2: Generating fake MRI to enhance Alzheimer’s disease prediction
All places booked
Author: Sebastian Gabriel Popescu, MPhil1
- Imperial College London, Great Britain
Abstract Alzheimer’s disease (AD), a common form of dementia, occurs most frequently in aged population. More than 30 million people worldwide suffer from AD and, due to the increasing life expectancy, this number is expected to triple by 2050. Because of the dramatic increase in the prevalence of AD, the identification of effective biomarkers for the early diagnosis and treatment of AD in individuals at high risk to develop the disease is crucial. Mild cognitive impairment (MCI) is a transitional stage between age-related cognitive decline and AD, and the earliest clinically detectable stage of progression towards dementia or AD. Despite of many efforts, identifying efficient AD-specific biomarkers for the early diagnosis and prediction of disease progression is still challenging and requires more research. Neuroimaging based algorithms using structural T1 scans have been used to predict conversion from MCI to AD (Moradi, 2015) but there is still room for improvement.
A major problem in classification of Alzheimer’s disease lies in the small datasets available. Hence, in this project our aim is to create synthetic/artificial T1 MRI scans and augment our dataset with the aim to further improve accuracy. Our workflow would involve a two-stage approach, whereby we would first generate fake MRI scans and secondly, we would add them to our existing real MRI scans and perform classification.
In terms of data augmentation techniques, Mixup (Zhang,2017) is a simple method which interpolates MRI scans originating from two different classes to obtain a new MRI scan with a fractional class label. Additionally, Generative Adversarial Networks (Goodfellow, 2014) have been successfully used to generate realistic pictures of celebrities or natural images with promising new developments in the realm of medical imaging (Wolterink, 2018).
Participants will be encouraged to develop their own classification pipeline, for example using Convolutional Neural Networks. Lastly, participants will be asked to consider ways of interpreting the predictions, to help better understand the neuroanatomical features involved in progression from MCI to AD.
A list of 1-5 key papers/materials summarising the subject:
- “Generative Adversarial Networks” by Goodfellow,2014
- “Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects” by Moradi et al.,2015
- “Mixup: Beyond Empirical Risk Minimization” by Zhang et al.,2017
- “Generative adversarial networks and adversarial methods in biomedical image analysis” by Wolterink et al.,2018
A list of requirements for taking part in the project:
- conversational English
- some experience in Machine Learning (basic understanding of CNNs and GANs)
- programming skills in Python, Tensorflow or PyTorch”
A maximal number of participants: 6
Skills and competences you can learn during the project:
- Understanding of the structural effects of Alzheimer’s disease on the brain
- Experience in classification using CNNs or more basic machine learning classifiers
- Data augmentation skills (Generative Adversarial Networks, MixUp)”
Is there a plan for extending this work to a paper in case the results are promising? Yes
Project 3: Network analysis of the electrical activity of the brain during epileptic seizures
All places booked
Authors: Jarosław Klamut, MSc 1 / Mateusz Wiliński, MSc 2
- Faculty of Physics, University of Warsaw, Poland
- Scuola Normale Superiore di Pisa, Italy
Abstract: The recent development in Network Science had a significant impact on many fields of science. Networks proved to be suitable tools in describing a variety of complex systems, brain being one of them. So-called brain networks, based mainly on fMRI or EEG signals, were characterized by a non-trivial hierarchical structure [1]. Interestingly, it was shown that neurological and psychiatric disorders are often accompanied by significant changes in this characteristic structure [2].
Despite initial successes, there is still a number of bottlenecks in the brain networks analysis. In particular, how to properly estimate connections between different parts of the brain? More specifically, how to get rid of bias imposed by volume conduction or active reference electrode? This bias, that affects all signal simultaneously is often referred to as the problem of common sources. It is widely accepted that neuronal synchrony is a good measure of functional integrity in the brain. There are, however, many ways to estimate it. Recent results suggest that methods based on Phase Lag Index are well suited when using EEG data. They were shown to successfully diminish bias from common sources and give better results both for synthetic and real data [3].
Epilepsy is a highly prevalent disorder. 1% of all emergency department visits in USA are caused by epilepsy. In 2004 it resulted in around $ 17 billion of economic costs in Europe, 2 billion in India and $ 1 billion in USA. Yet the brain activity during seizures is still not well explained in the scientific literature. Its connection to brain electrical activity makes it natural to choose EEG as an experimental tool to collect the data suitable for diagnosis. In the project, we will provide with EEG datasets from patients with epilepsy and we will attempt to determine whether the brain network structure changes during epileptic seizures. In order to approach this question, we will use Phase Lag Index based measures. Finally, we would like to determine whether the network characteristics may be more successful in distinguishing or even predicting, different epileptic states, in comparison with classical indicators, such as the signal variance.

A list of 1-6 key papers/materials summarising the subject:
- Meunier, D., Lambiotte, R., & Bullmore, E. T. (2010). Modular and hierarchically modular organization of brain networks. Frontiers in neuroscience, 4, 200.
- Stam, C. J. (2014). Modern network science of neurological disorders. Nature Reviews Neuroscience, 15(10), 683.
- Stam, C. J., Nolte, G., & Daffertshofer, A. (2007). Phase lag index: assessment of functional connectivity from multi channel EEG and MEG with diminished bias from common sources. Human brain mapping, 28(11), 1178-1193.
- Bullmore, Ed, and Olaf Sporns. “Complex brain networks: graph theoretical analysis of structural and functional systems.” Nature Reviews Neuroscience 10.3 (2009): 186.
- https://en.wikipedia.org/wiki/Epilepsy and https://en.wikipedia.org/wiki/Electroencephalography
- Scheffer, Marten, et al. “Early-warning signals for critical transitions.” Nature 461.7260 (2009): 53.
List of useful skills in the project:
- Neurobiological knowledge
- Signal analysis
- Data visualization
- EEG signal analysis and modelling
- Network analysis
- Programming in Python, Igraph (python library)
- English (reading, writing)
- BSc program, or higher
- Bring computer
A maximal number of participants: 6
Skills and competences to be acquired during the project:
- Team work (programming and solving problems in parallel)
- Presenting scientific results (writing paragraphs, creating figures etc.)
- Data preprocessing
Is there a plan for extending this work to a paper in case the results are promising? Not sure
Project 4: Deep Frankenstein: dissecting and sewing artificial neural networks
All places booked
Authors: Piotr Migdał, PhD1 / Katarzyna Kańska 2
- p.migdal.pl
- Faculty of Mathematics, Informatics, and Mechanics, University of Warsaw, Poland
Abstract: Convolutional neural networks are the state-of-the-art solution for image classification. They are able to recognize images at near-human (or occasionally: super-human) level. Their basic units are very different from the biological ones. Each layer is an array, for processing we use convolutions and matrix multiplications, and for gating - simple mathematical functions. Yet, the global architecture of local, hierarchical processing was inspired by mammalian visual cortex.
In this project we are going to explore which kind of information is being processed. We will try dissecting a network to see if a subset of network is able to recognize an image. We will try to see if different neural ImageNet architecture (e.g. VGG, ResNet, Inception, etc) “see” similar features.
And, in the absence of any ethics commission for artificial neural networks, we will cut two networks, sew them, and see is this frankensteinian model is able to recognize images.
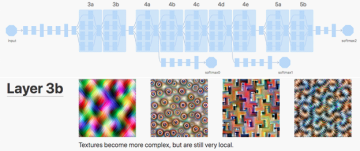
A list of 1-5 key papers/materials summarising the subject:
- Chris Olah et al., “Feature Visualization”, Distill, https://distill.pub/2017/feature-visualization/
- Jason Yosinski et al., “How transferable are features in deep neural networks?”, https://arxiv.org/abs/1411.1792
- Radoslaw Martin Cichy et al., “Comparison of deep neural networks to spatio-temporal cortical dynamics of human visual object recognition reveals hierarchical correspondence”, https://www.nature.com/articles/srep27755
- Daniel L. K. Yamins et al., “Performance-optimized hierarchical models predict neural responses in higher visual cortex”, https://www.pnas.org/content/111/23/8619
List of useful skills in the project:
- Expert knowledge related to the visual cortex
- Psychology of vision
- Academic and technical writing
- Data visualization (academic, technical or artistic)
- I assume at least basic experience with programming and Python. If not yet, I gathered some materials there: https://p.migdal.pl/2016/03/15/data-science-intro-for-math-phys-background.html
- To get overview of deep learning for image classification, I suggest reading my blog post: https://p.migdal.pl/2017/04/30/teaching-deep-learning.html
- Please, read and run code (on Kaggle, Neptune or your local computer) for: https://deepsense.ai/keras-vs-pytorch-avp-transfer-learning/
- I really encourage you to try transfer learning (in either Keras or PyTorch) for a different dataset (pick any, e.g. chihuahua or muffin) and send screenshots of a few classified, and misclassified examples (along with their probabilities). In case there is more people interested in this project than spots, I will use it as a qualification criterion.
- If you have any questions, feel invited to email me!
Skills and competences to be acquired during the project:
- Insight into the inner workings of deep learning (artificial neural networks) and analogies with biological image processing
A maximal number of participants: 10
Is there a plan for extending this work to a paper in case the results are promising? yes
Project 5: Tackling the ABCD Neurocognitive Prediction Challenge
All places booked
Authors: Hanna Nowicka 1
1.Oxford Centre for Functional MRI of the Brain (FMRIB), Wellcome Centre for Integrative Neuroimaging (WIN), University of Oxford, United Kingdom
Abstract: The objective of the project is to propose the method for predicting fluid intelligence in children from the T1-weighted MRI images. Work will most likely consist of data preprocessing, dimensionality reduction and fitting a regression model. First idea will be to apply methods like PCA for exploratory analysis and VAEs for nonlinear dimensionality reduction. Then we will use the space from VAEs to gaussian process regression and test different kernels. Of course, other approaches are also possible, depending on the preliminary results and the interest of the team. If successful, the aim is to submit the method to the ABCD Neurocognitive Prediction Challenge which is hold in conjugation with MICCAI 2019 conference. Preprocessed data is provided by the challenge organisers (requires registration in advance). More details about the problem and the data are available at: https://sibis.sri.com/abcd-np-challenge/.
A list of 1-5 key papers/materials summarising the subject:
- Challenge website: https://sibis.sri.com/abcd-np-challenge/ (information about the challenge and how to register to access the data)
- Details about the data processing performed: https://www.biorxiv.org/content/early/2018/11/04/457739
- ‘Introduction to Neuroimaging Analysis’ – Oxford Neuroimaging Primers, Mark Jenkinson and Michael Chappell, Oxford University Press, 2017
- Some read about fluid intelligence
A list of requirements for taking part in the project:
- IMPORTANT: Registration to access the data prior to the hackathon is strictly required. Sadly, we cannot allow anyone without the DUC approval to work with the data (and it might take few days to obtain the access as it requires the institution support) so please do that in advance, if interested in taking part in the project!
- English skills good enough to communicate with international team.
- At least basic Python skills.
A maximal number of participants on the project: 6
Skills and competences you can learn during the project: The main gain from the project would be having a submission to the MICCAI challenge. Hopefully, we can get group with varied skills and learn from each other skills like: basics of structural brain MRI processing and interpretation, data science and visualisation, machine learning in Python.
Is there a plan for extending this work to a paper in case the results are promising? Not sure
Project 6: Multidimensionality problem in passive BCI for game control
Authors: Maciej Rudziński 1 /Adrian Elczewski
1.BrainAttach; Brain Tracking
Abstract: In BCI, the environment is controlled by users through listening to active mental commands that need active effort and learning process. In this project, we are proposing a different paradigm where BCI algorithms listen to well-defined passive bottom-up processes like emotions and attention to choose/guess user preferred action. This approach is similar to Flying Mollusk work but we are proposing its further development and more specific emotions indicators as a starting point. We are providing
- 2 EEG setups with 2 channels (5 electrodes) and 1k Hz sampling
- Web service that:
- Cleans signal and removes artifacts
- Converts signal into emotions (BIS-BAS*) and attention indicators
- Is integrated to the provided game for ease of work
- Short lecture about BIS-BAS
- Short lecture on signal quality and cleaning methods
- Introduction to the problem of multidimensionality and timescales, in the use of emotions, for controlling (or assisting) games and devices
- Presentation of sample Python game reacting to emotions (cannon) and how to use indicator e.g shooting on BAS
- Walk through Python game code http://www.grantjenks.com/docs/freegames/
- Help with development, expanding and optimizing parameters (The “problem of multidimensionality and timescales”)
- Brains tournament in cannon between teams (if time permits)
A list of 1-5 key papers summarising the subject:
- http://gamestudies.org/1801/articles/david_melhart
- http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.96.352&rep=rep1&type=pdf
- https://www.ted.com/talks/mihaly_csikszentmihalyi_on_flow?language=en
A list of requirements for taking part in the project:
- English : communicative,
- Python experience above 1 month of active use,
- Psychology basic concepts (emotions) would be a plus,
- ML experience will give you more fun
A maximum number of participants: 10
What can the participant gain from the project?
- Transferring/joining knowledge from neuroscience/psychology with practical application in BCI.
- How to create player aware games.
- Interpretation of BIS-BAS and Flow concept in real life situation.
- How attention is linked to emotions.
Is there a plan for extending this work to a peer-reviewed paper in case the results are promising? Not sure
Project 7: Cross-Recurrence Quantification Analysis (cRQA) as a new measure of functional connectivity
All places booked
Authors: Krzysztof Bielski 1 / Joanna Rączaszek-Leonardi 2
- Nencki Institute of Experimental Biology PAS, Warsaw, Poland
- Faculty of Psychology, University of Warsaw, Poland
Abstract: The human brain organization might be characterized by two basic properties: the segregation and the integration (Friston, 1994). While the functional segregation describes specialization of the brain areas or networks, the functional integration provides an insight on how information between specialized areas/networks is integrated. This second property is frequently studied by observing brain connectivity patterns. In recent years, one type of connectivity namely functional connectivity has gained a great attention. In fact, actually this concept is not directly associated with biologically-rooted connections (for example, long-range white matter tracts) but rather with the similarity between time-series of BOLD, EEG or other neuroimaging signals. It follows logic: if signals from two brain areas are similar, these two brain regions are involved in processing similar information and thus, they are functionally connected. Most often, such functional connections are assessed with measures like the Pearson correlation. However, relatively smaller number of studies deal with measures directly associated with temporal structures in the data (e.g. mutual information etc.). In our project, we would like to develop a method introduced by Lombardi et al. (2017) for estimating functional connectivity patterns known from the Dynamical Systems Theory - cross Recurrence Quantification Analysis (cRQA). cRQA is a method which find application in analyzing coupling dynamical systems such as social interactions in dyads, interdependence of EEG signals etc. It allows presenting complex interactions in the simple form of a vector of measures describing state-switching properties of time-series. Following observations by Lombardi et al. (2017), we aim to compare obtained connectivity patterns defined by the cRQA with connectivity patterns obtained with the Pearson Correlation. We will use task-fMRI data from procedures testing different cognitive and emotional processes as well as resting-state fMRI data taken from the Human Connectome Project database.
A list of 1-5 key papers summarising the subject:
- Lombardi, A., Tangaro, S., Bellotti, R., Bertolino, A., Blasi, G., Pergola, G., … Guaragnella, C. (2017). A Novel Synchronization-Based Approach for Functional Connectivity Analysis. Complexity, 2017, 1–12. https://doi.org/10.1155/2017/7190758
- Marwan, N., & Webber, C. L. (2015). Mathematical and Computational Foundations of Recurrence Quantifications. In C. L. Webber, & N. Marwan (Eds.), Recurrence Quantification Analysis: Theory and Best Practices. Cham: Springer International Publishing. https://doi.org/10.1007/978-3-319-07155-8
- Bastos, A. M., & Schoffelen, J.-M. (2016). A Tutorial Review of Functional Connectivity Analysis Methods and Their Interpretational Pitfalls. Frontiers in Systems Neuroscience, 9(January), 1–23. https://doi.org/10.3389/fnsys.2015.00175
A list of requirements for taking part in the project:
- Students from Bachelor degree level or higher
- Communicative English
- Basic programming skills in Python and Matlab
Maximum number of participants: 6
What can the participant gain from the project?
- Cooperation in small team
- Knowledge about applying Dynamical Systems Theory to neuroimaging data
- Knowledge about analysis of functional connectivity
Is there a plan for extending this work to a peer-reviewed paper in case the results are promising? Yes
Project 8: EMG-based desktop controlling
Authors: Paweł Pierzchlewicz 1
- University of Warsaw, Faculty of Physics, Poland
Abstract: The daily usage of our computers can become daunting at times: we spend our days slouched by the keyboard. What if we could create a method for controlling the computer using gestures? The idea is to create a prototype for a wearable Human-Computer Interface (HCI) based on electromyography (EMG) signals. We will start off by gathering training data. Later through the use of Barachant’s Minimum Distance to Mean (MDM) (https://hal.archives-ouvertes.fr/file/index/docid/681328/filename/Barachant_tbme_final.pdf) algorithm we will attempt to classify some simple gestures. This will be handled by a raspberry pi if possible. Finally, a client on a target remote computer will capture the determined gestures and map them into useful actions like opening the browser etc.
This will require certain tasks to be performed: gathering of learning data, creating a classification model, building a client app to map the classified action.
A list of 1-5 key papers / online materials summarising the subject:
- https://hal.archives-ouvertes.fr/file/index/docid/681328/filename/Barachant_tbme_final.pdf,
- https://ieeexplore.ieee.org/abstract/document/1570513
List of useful skills in the project:
- Graphic design,
- Muscle anatomy,
- Motor neuron physiology,
- Modelnig,
- Comfortable with python,
- English good enough to read code documentation and some research articles,
- Signal processing would be a plus
Maximum number of participants: 10
What participants gain/learn from this project: They can learn an interesting method for signal analysis. Can gain experience with biosignal experimental setups.
Is there a plan for extending this work to a peer-reviewed paper in case the results are promising? Yes
Project 9: Mapping the state of modern psychology through the public online activity of scientists
Authors: Daniel Borek 1
- Department of Data Analysis, Faculty of Psychology and Educational Sciences, Ghent University, Ghent, Belgium
Abstract: Many active scientists use online social networks to share their research. Many of the ideas shaping modern science are disseminated this way. Online discussions between scientists precede and often shape what is published in journals.
This project aims to map relations and mutual influence in online science communities. Psychology and neuroscience communities are especially interesting. These fields are accompanied by transient changes, due to replication crisis, open science movement and many methodological debates and advances.
Is it possible to map the structure of the field and “hot” topics using mostly Twitter data? How the information about publications spreads? What define that the paper will become popular (apart from scientific merit)? These questions are relevant not only for disciplines such as sociology of science. Also, answers could inform those, who want to share their research with others in the most effective way. P
The project could use existing dataset. We will also download relevant data directly from Twitter to answer specific problems. Please remember that there are some limitations imposed on downloading data from the service. We will process and analyze data using open languages like R or Python. We could use Gephi for visualizations. Example analysis could involve simple graph-theoretical measures to map the relation between different sub-disciplines.
Further direction for the analysis will depend on participants and the availability of data. The project description soon will be accompanied by a small demo. Other ideas on how to analyze the public online presence of scientists are also welcome.
A list of 1-6 key papers / online materials summarising the subject:
- Example of mapping the field structure using published papers https://homepage.univie.ac.at/noichlm94/posts/structure-of-recent-philosophy-ii/
- Example of mapping the networks of economist on twitter
- https://io.mongeau.net/repec-twitter-network/?fbclid=IwAR0Z07I_3FmGjTofeXa1omEACdxdcfHlxUE44J-W_yjLx7WWFBCvXMXUDjs
- Downloading tweets: http://social-metrics.org/downloading-tweets-by-a-list-of-users-take2/
- Similar analyses in humanities: http://journalofdigitalhumanities.org/1-3/the-impact-of-social-media-on-the-dissemination-of-research-by-melissa-terras/ https://arxiv.org/pdf/1807.05571.pdf
- How to make a graph of twitter followers: https://github.com/alumbreras/twitter-followers-graph
List of useful skills in the project:
- BSc program, or higher
- English: good, not necessarily proficient
- Good Python/R programming skills and basic familiarity with machine learning is a plus, but you could participate even when you cant program
Maximum number of participants: 12
Is there a plan for extending this work to a peer-reviewed paper in case the results are promising? Yes
Preliminary schedule
|
Friday, 1st March 2019 |
Saturday, 2nd March 2019 |
Sunday, 3rd March 2019 |
|
|
9:00 |
Brain hacking |
Brain hacking |
|
|
10:00 |
Brain hacking |
Brain hacking |
|
|
11:00 |
Brain hacking |
Brain hacking |
|
|
12:00 |
Lunch |
Brain hacking |
|
|
13:00 |
Brain hacking |
Lunch |
|
|
14:00 |
Brain hacking |
Preparing final presentations |
|
|
15:00 |
Talks from: |
A round of 10-min final presentations |
|
|
16:00 |
Tea-time break |
||
|
17:00 |
Opening, welcome drinks |
Brain hacking |
Goodbye drinks |
|
18:00 |
5-min blitz project opening presentations |
Brain hacking |
|
|
19:00 |
Break for further drinks |
Dinner |
|
|
19:30 |
Ignite talk: Roberto Toro |
||
|
20:00 |
Brain hacking |
||
|
20:30 |
Brainstorming |
||
|
21:00 |
Brain hacking |
||
|
21:30 |
Late-night social |
||
|
22:00 |
Brain hacking |
||
|
23:00 |
Brain hacking |
Participant Registration
Participant Registration is now CLOSED.
Registration will take place in three rounds and there will be small registration fee (to cover the catering during the event) for the project participants:
- EARLY -> 21.01 - 03.02 -> 100 PLN / 25 EUR
- REGULAR -> 04.02 - 17.02 -> 120 PLN / 29 EUR
- LATE -> 18.02 - 24.02 -> 135 PLN / 33 EUR
PLEASE NOTE: the first round is paid after qualifying, second and third immediately
If you have any questions or queries about fees, please write to us: brainhackwarsaw@gmail.com
Payment details
- Bank account: Bank Millennium 44 1160 2202 0000 0000 6084 9470
- IBAN: PL 44 1160 2202 0000 0000 6084 9470
- SWIFT/BIC: BIGBPLPW
- Transfer title: Brainhack Warsaw 2019
- If you want to get an invoice, please send us the necessary information to: invoice.brainhack@gmail.com
For your convenience we prepared a list of nice and friendly hotels where you get a special Brainhack Warsaw discount:
- Moon Hostel - 10% discount but remember to rely on participation in Brainhack
- Lwowska Hostel - reservaation only by rezerwacja@hostellwowska11.pl, for 4-person room you get 55 PLN per person and 50 PLN per person in 6-person room on code: Brainhack
- Mish Mash Hostel - reservation by mishmashhostel.com, for 1-3.03 and code MMBrainhack2019 you get 10% discount
- Tatamka Hostel - 10% discount on phone reservations with the password of brainhack
- Soundgarden Hotel - on 1-3.03, a normal room without breakfast costs 190 PLN per night, for the password: brainhack2019. And here is the instruction how to make a reservation using the code.




